MRNet: Deep-learning-assisted diagnosis for knee magnetic resonance imaging
Nicholas Bien *, Pranav Rajpurkar *, Robyn L. Ball, Jeremy Irvin, Allison Park, Erik Jones, Michael Bereket, Bhavik N. Patel, Kristen W. Yeom, Katie Shpanskaya, Safwan Halabi, Evan Zucker, Gary Fanton, Derek F. Amanatullah, Christopher F. Beaulieu, Geoffrey M. Riley, Russell J. Stewart, Francis G. Blankenberg, David B. Larson, Ricky H. Jones, Curtis P. Langlotz, Andrew Y. Ng†, Matthew P. Lungren†
We developed an algorithm to predict abnormalities in knee MRI exams, and measured the clinical utility of providing the algorithm’s predictions to radiologists and surgeons during interpretation.
Magnetic resonance (MR) imaging of the knee is the standard of care imaging modality to evaluate knee disorders, and more musculoskeletal MR examinations are performed on the knee than on any other region of the body. In a study published in PLOS medicine, we developed a deep learning model for detecting general abnormalities and specific diagnoses (anterior cruciate ligament [ACL] tears and meniscal tears) on knee MRI exams. We then measured the clinical utility of providing the model’s predictions to clinical experts during interpretation.
Consider this knee MR exam, shown (top row) in the three series — can you find the abnormality (click images to pause)? The deep learning algorithm is able to identify the ACL tear (best seen on the sagittal series) and localize the abnormalities (bottom row) using a heat map which displays increased color intensity where there is most evidence of abnormalities.
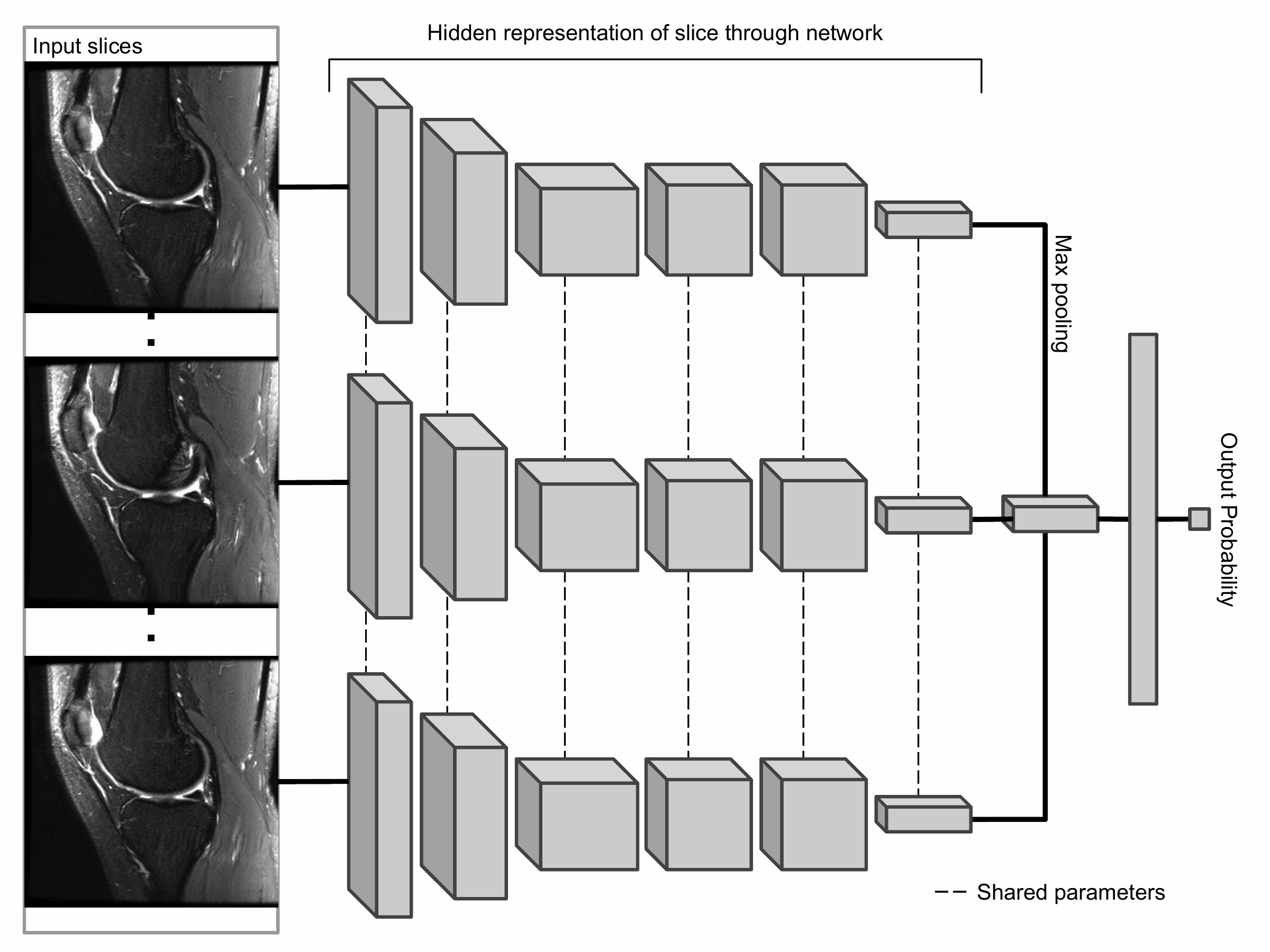
The primary building block of our prediction system is MRNet, a convolutional neural network (CNN) mapping a 3-dimensional MRI series to a probability.
The input to MRNet has dimensions s × 3 × 256 × 256, where s is the number of images in the MRI series (3 is the number of color channels). First, each 2-dimensional MRI image slice is passed through a feature extractor to obtain a s × 256 × 7 × 7 tensor containing features for each slice. A global average pooling layer is then applied to reduce these features to s × 256. We then applied max pooling across slices to obtain a 256-dimensional vector, which is passed to a fully connected layer to obtain a prediction probability.
Because MRNet generates a prediction for each of the sagittal T2, coronal T1, and axial PD series, we train a logistic regression to weight the predictions from the 3 series and generate a single output for each exam.
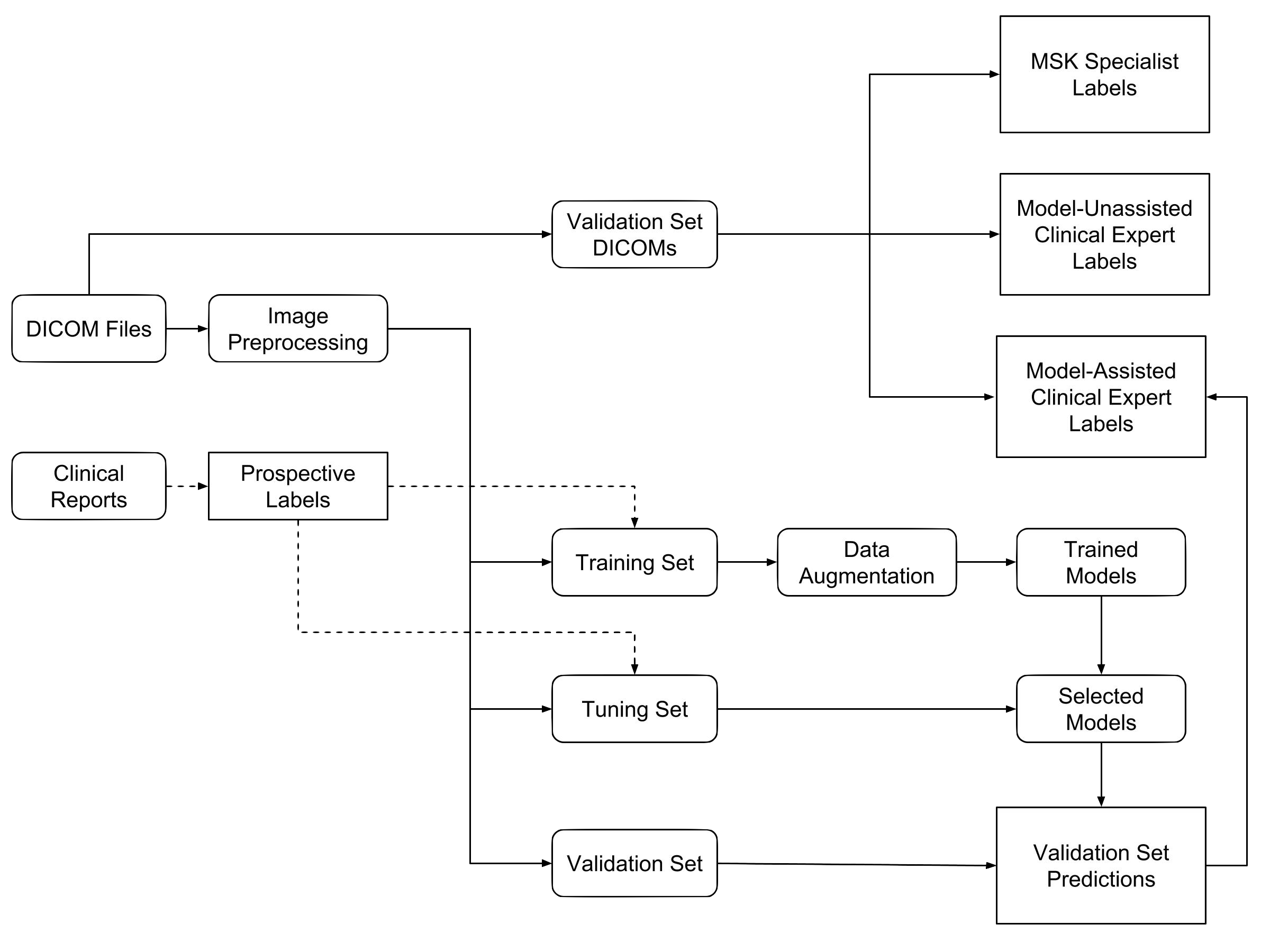
We measured the performance of general radiologists and surgeons with and without model assistance.
7 practicing board-certified general radiologists and 2 practicing orthopedic surgeons at Stanford University Medical Center (3–29 years in practice, average 12 years) read a validation set of 120 exams twice, once without model assistance and once with model assistance, separated by a washout period of at least 10 days. For the reads with model assistance, model predictions were provided as predicted probabilities of a positive diagnosis for each of the 3 labels (e.g., 0.98 ACL tear, 0.7 Meniscal tear, and 0.99 abnormal).
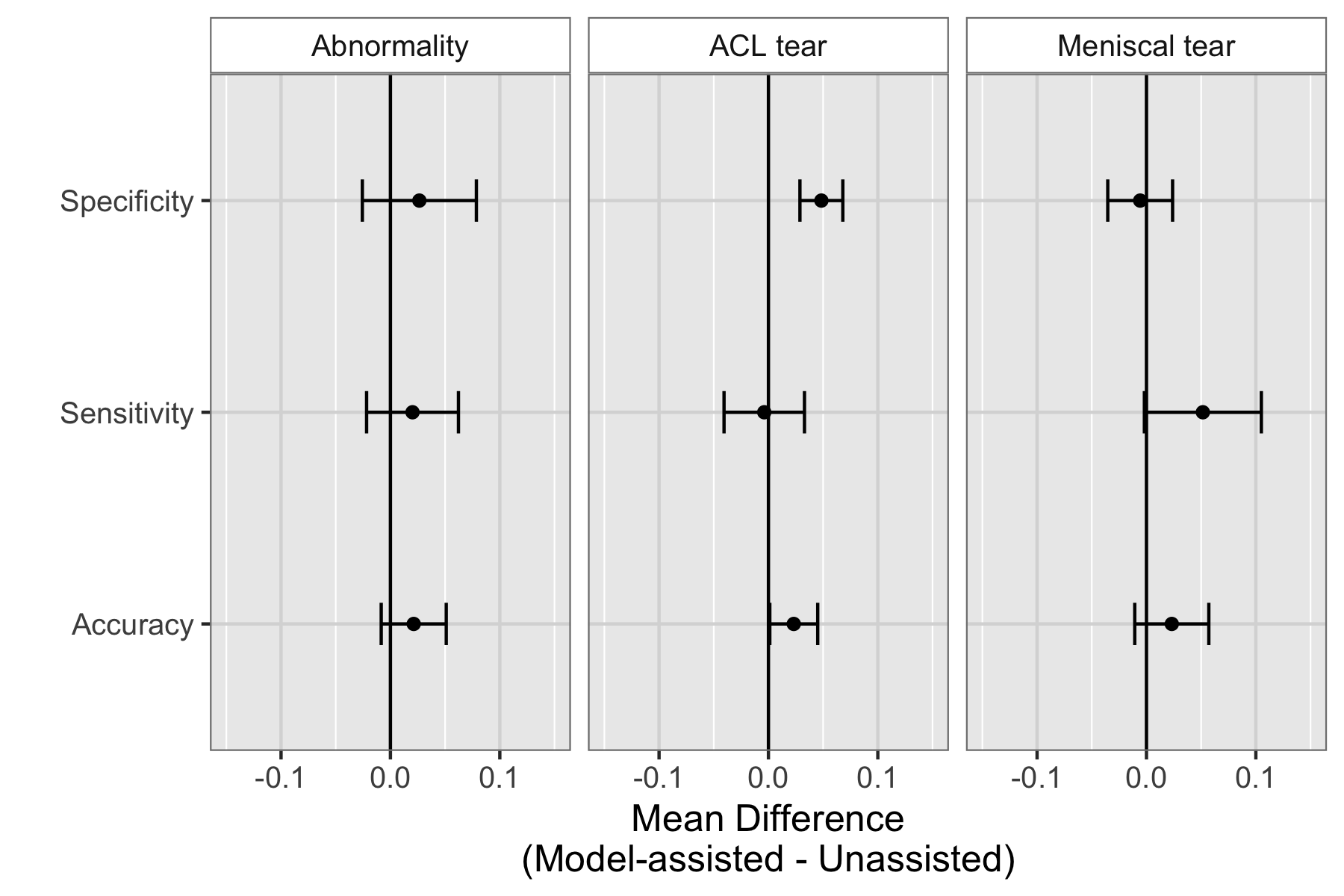
We found that model assistance significantly reduced the rate at which healthy patients would be mistakenly diagnosed as having ACL tears.
Model assistance resulted in a mean increase of 0.048 (4.8%) in ACL specificity: for every 100 healthy patients, ~5 are saved from being unneccessarily considered for surgery.
Though it appeared that model assistance also significantly increased the clinical experts’ accuracy in detecting ACL tears and sensitivity in detecting meniscus tears, these findings were no longer significant after adjusting for multiple comparisons by controlling the False Discovery Rate.
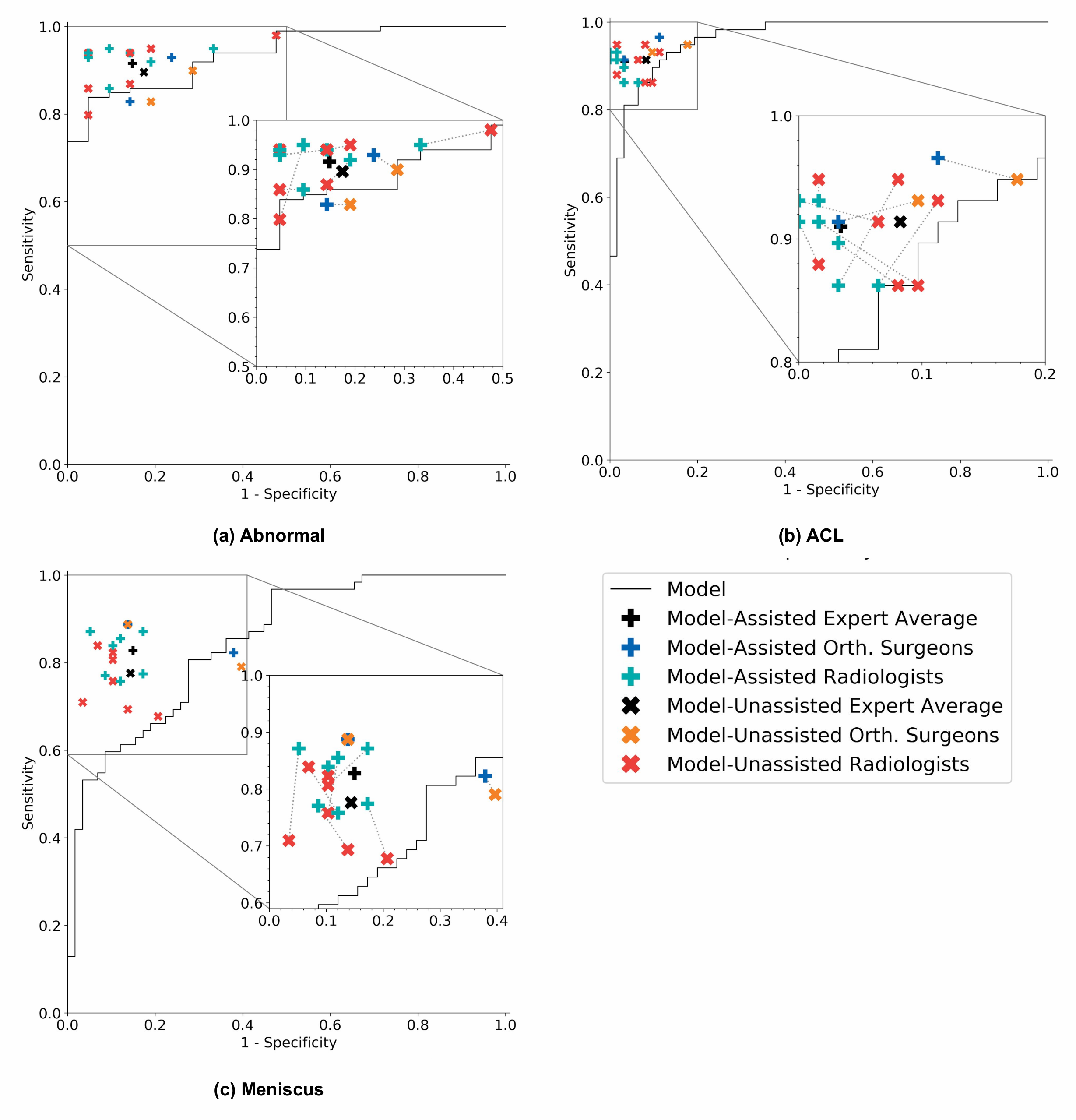
On its own, the model achieved AUCs of 0.937 (95% CI 0.895, 0.980), 0.965 (95% CI 0.938, 0.993), and 0.847 (95% CI 0.780, 0.914) for abnormality detection, ACL tear detection, and meniscal tear detection respectively. Notably, the model achieved high specificity in detecting ACL tears on the internal validation set, which suggests that such a model, if used in the clinical workflow, may have the potential to effectively rule out ACL tears. We also compared the performance of the model to that of general radiologists (unassisted), and found that in detecting abnormalities, there were no significant differences in the performance metrics of the model and general radiologists. In detecting ACL tears, general radiologists achieved significantly higher sensitivity than the model, and in detecting meniscal tears achieved significantly higher specificity.
We validated MRNet on a dataset from a different institution, and found that the model picked up ACL tears with high discriminative ability.
We obtained a public dataset of 917 exams with sagittal T1-weighted series and labels for ACL injury from Clinical Hospital Centre Rijeka, Croatia. On the external validation set of 183 exams, the MRNet trained on Stanford sagittal T2-weighted series achieved an AUC of 0.824 (95% CI 0.757, 0.892) in the detection of ACL injuries with no additional training; an MRNet trained on the rest of the external data achieved an AUC of 0.911 (95% CI 0.864, 0.958).

We are excited to partner with healthcare providers that are interested in working with us to further validate AI technologies in medical imaging.
Automated abnormality prediction and localization could help general radiologists or even non-radiologist clinicians (such as orthopedic surgeons) interpret medical imaging for patients at the point of care rather than waiting for specialized radiologist interpretation, which could aid in efficient interpretation, reduce errors, and help standardize quality of diagnoses when specialist radiologists are not readily available.
Ultimately, more studies are necessary to evaluate the optimal integration of this model and other deep learning models in the clinical setting. We hope to partner with healthcare providers to research and validate automatic AI models in medical imaging.
Source: MRNet: Deep-learning-assisted diagnosis for knee magnetic resonance imaging